


 下载产品说明书
下载产品说明书 用小程序,查商品更便捷
用小程序,查商品更便捷



 收藏
收藏
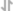 对比
对比 咨询
咨询
Product Usage Information
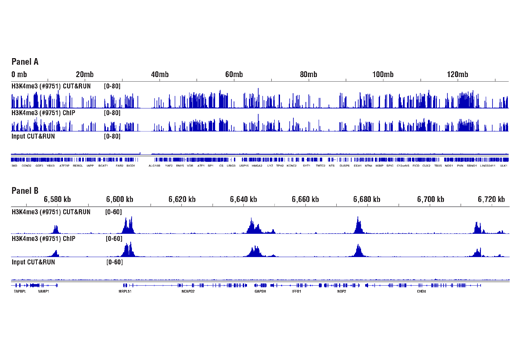
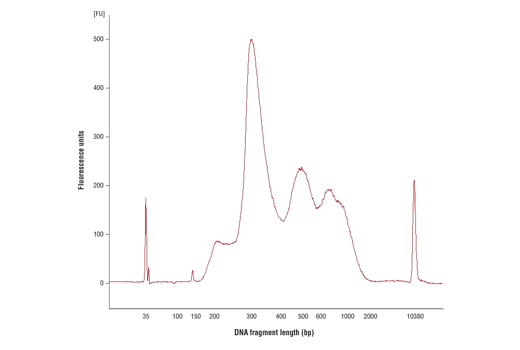
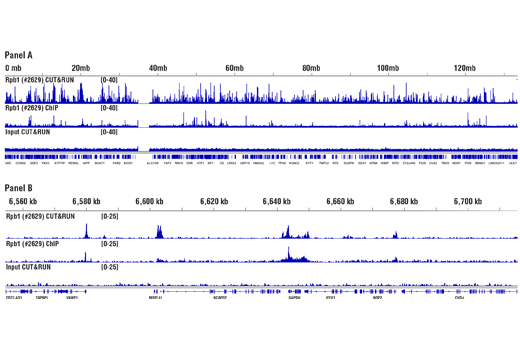
For the pAG-MNase Enzyme, after cell permeabilization and primary antibody binding, resuspend cells in 50 μl of digitonin buffer containing 1.5 μl of pAG-MNase Enzyme (33X dilution). Incubate cell samples with rotation at 4°C for 1 hour, wash cells with digitonin buffer, and then perform the chromatin digestion. Sample Normalization Spike-In DNA can be added directly to the digestion stop buffer. For sample normalization with NG-seq, add 5 μl (50 pg) of Sample Normalization Spike-In DNA to each reaction. When using 100,000 cells or 1mg of tissue per reaction this ensures that the normalization reads are around 0.5% of the total sequencing reads. If more or less than 100,000 cells or 1mg of tissue are used per reaction, proportionally scale the volume of Sample Normalization Spike-In DNA up or down to adjust normalization reads to around 0.5% of total reads. When performing sample normalization, be sure to map the CUT&RUN sequencing data for all samples to both the test reference genome (e.g. human) and the sample normalization genome (S. cerevisiae).







 危险品化学品经营许可证(不带存储) 许可证编号:沪(杨)应急管危经许[2022]202944(QY)
危险品化学品经营许可证(不带存储) 许可证编号:沪(杨)应急管危经许[2022]202944(QY)  营业执照(三证合一)
营业执照(三证合一)