


 下载产品说明书
下载产品说明书 用小程序,查商品更便捷
用小程序,查商品更便捷



 收藏
收藏
 对比
对比 咨询
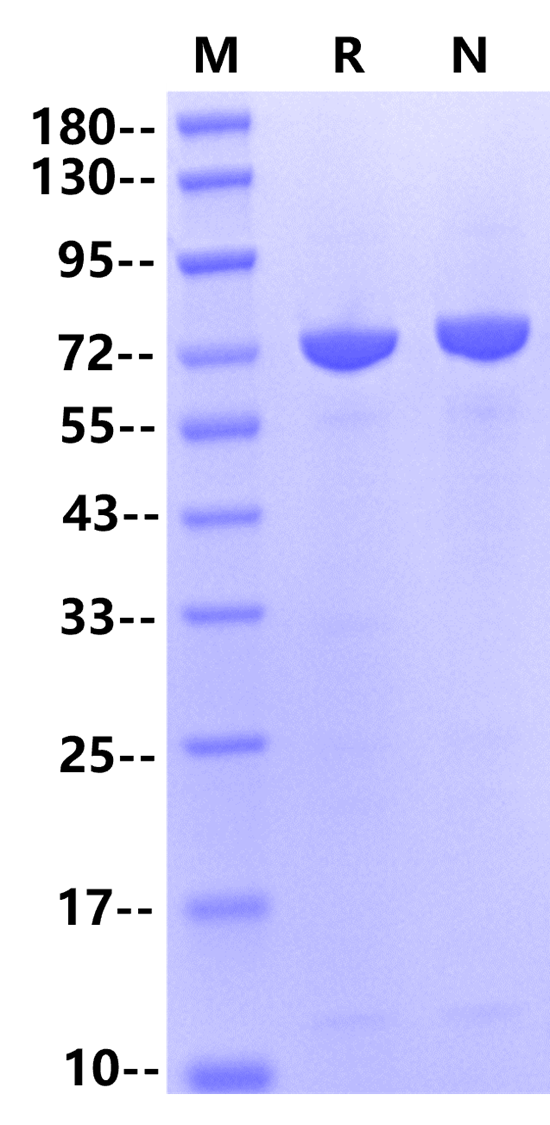
咨询72kDa (Reducing)
72kDa (Reducing)

72kDa (Reducing)







Storage Solution: 2 U/ul DnaseⅠ、10mM Tris-Hcl、2mM CaCl2、50%Glycerol (pH7.6, 25℃)
10*Reaction
Buffer: 100mM Tris-Hcl、25mM MgCl2、5mM CaCl2 (pH7.6, 25℃)



1μg (R: reducing condition, N: non-reducing\ncondition).
"}]

DNase I
(Deoxyribonuclease I), can digest single or double-stranded DNA to produce mono
deoxynucleotides or single or double-stranded oligo deoxynucleotides, its
optimal working pH range is 7-8. DNase I activity is dependent on Ca2+
and can be activated by other bivalent metal ions such as Mg2+, Mn2+,
Zn2+, etc. In the presence of Mg2+, the enzyme can
randomly recognize and cut any site on any strand of DNA. In the presence of Mn2+,
two strands of DNA can be cut at the same site to form sticky ends with flat
ends or 1-2 nucleotides protruding.

Store at -25 ~ -15℃
for 2 years
[1] Vanecko S, Laskowski M. Studies of the Specificity of Deoxyribonuclease I[J]. Journal of Biological Chemistry, 1961, 236(236):3312-6.
[2] Kienzle N, Young D, Zehntner S, et al. DNaseI treatment is a prerequisite for the amplification of cDNA from episomal-based genes[J]. Biotechniques, 1996, 20(4):612-6.
[3] Michael,R, Green, et
al. Human β-globin pre-mRNA synthesized in vitro is accurately spliced in
xenopus oocyte nuclei[J].Cell, 1983, 32(3):681-694.

参考图片
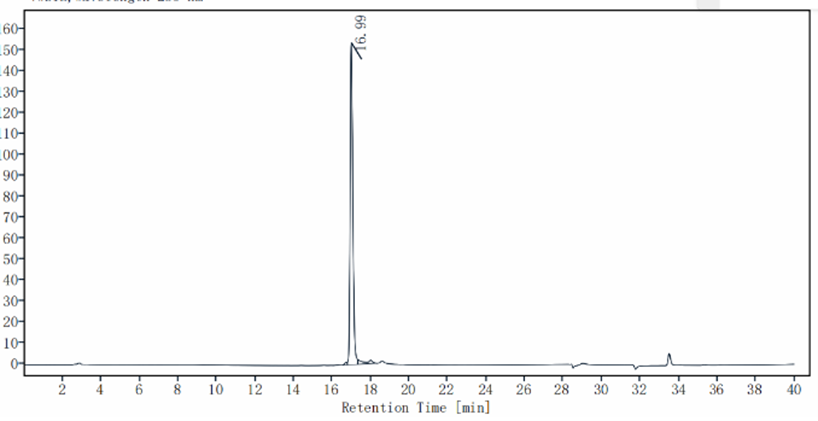
1μg (R: reducing condition, N: non-reducing condition).
The results of 1μg pBR322 plasmid digestion separated under different quantity of DnaseⅠ, The reaction was incubated for 10 minutes at 37°C, and 1% agarose gel was used for electrophoresis analysis after reaction.
M, marker;
Lane 1 1μg pBR322;
Lane 2 1μg pBR322 add 4U DNase I
Lane 3 1μg pBR322 add 2U DNase I
Lane 4 1μg pBR322 add 1U DNase I
Lane 5 1μg pBR322 add 0.5U DNase I
Lane 6 1μg pBR322 add 0.25U DNase I
Lane
7 1μg pBR322 add 0.125U DNase I





 危险品化学品经营许可证(不带存储) 许可证编号:沪(杨)应急管危经许[2022]202944(QY)
危险品化学品经营许可证(不带存储) 许可证编号:沪(杨)应急管危经许[2022]202944(QY)  营业执照(三证合一)
营业执照(三证合一)