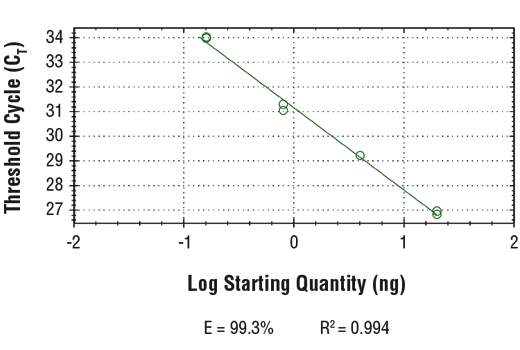
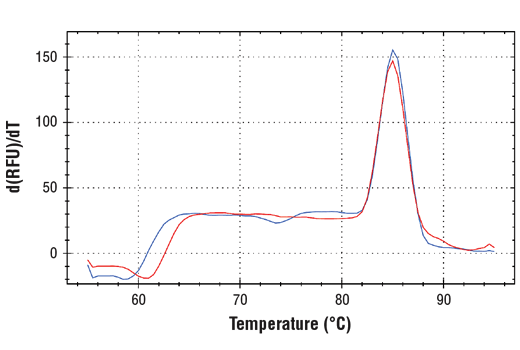
The chromatin immunoprecipitation (ChIP) assay is a powerful and versatile technique used for probing protein-DNA interactions within the natural chromatin context of the cell (1,2). This assay can be used to either identify multiple proteins associated with a specific region of the genome or to identify the many regions of the genome bound by a particular protein (3-6). ChIP can be used to determine the specific order of recruitment of various proteins to a gene promoter or to "measure" the relative amount of a particular histone modification across an entire gene locus (3,4). In addition to histone proteins, the ChIP assay can be used to analyze binding of transcription factors and co-factors, DNA replication factors, and DNA repair proteins. When performing the ChIP assay, cells are first fixed with formaldehyde, a reversible protein-DNA cross-linking agent that "preserves" the protein-DNA interactions occurring in the cell (1,2). Cells are lysed and chromatin is harvested and fragmented using either sonication or enzymatic digestion. Fragmented chromatin is then immunoprecipitated with antibodies specific to a particular protein or histone modification. Any DNA sequences that are associated with the protein or histone modification of interest will co-precipitate as part of the cross-linked chromatin complex and the relative amount of that DNA sequence will be enriched by the immunoselection process. After immunoprecipitation, the protein-DNA cross-links are reversed and the DNA is purified. Standard PCR or quantitative real-time PCR are often used to measure the amount of enrichment of a particular DNA sequence by a protein-specific immunoprecipitation (1,2). Alternatively, the ChIP assay can be combined with genomic tiling micro-array (ChIP on chip) techniques, high throughput sequencing (ChIP-Seq), or cloning strategies, all of which allow for genome-wide analysis of protein-DNA interactions and histone modifications (5-8). SimpleChIP® primers have been optimized for amplification of ChIP-isolated DNA using real-time quantitative PCR and provide important positive and negative controls that can be used to confirm a successful ChIP experiment.
1.Orlando, V. (2000) Trends Biochem Sci 25, 99-104.
2.Kuo, M.H. and Allis, C.D. (1999) Methods 19, 425-33.
3.Agalioti, T. et al. (2000) Cell 103, 667-78.
4.Soutoglou, E. and Talianidis, I. (2002) Science 295, 1901-4.
5.Mikkelsen, T.S. et al. (2007) Nature 448, 553-60.
6.Lee, T.I. et al. (2006) Cell 125, 301-13.
7.Weinmann, A.S. and Farnham, P.J. (2002) Methods 26, 37-47.
8.Wells, J. and Farnham, P.J. (2002) Methods 26, 48-56.
















 用小程序,查商品更便捷
用小程序,查商品更便捷






 危险品化学品经营许可证(不带存储) 许可证编号:沪(杨)应急管危经许[2022]202944(QY)
危险品化学品经营许可证(不带存储) 许可证编号:沪(杨)应急管危经许[2022]202944(QY)  营业执照(三证合一)
营业执照(三证合一)