 全部商品分类
全部商品分类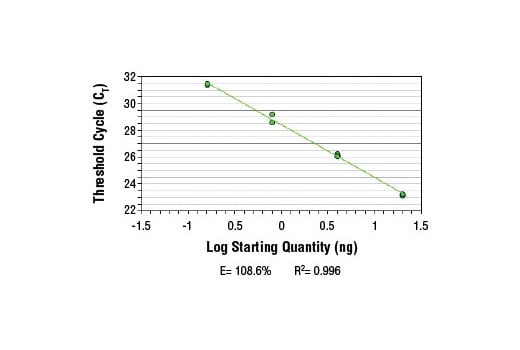


SimpleChIP® Human Tyrosinase Promoter Primers contain a mix of forward and reverse PCR primers that are specific to a region of the human tyrosinase promoter. These primers can be used to amplify DNA that has been isolated using chromatin immunoprecipitation (ChIP). Primers have been optimized for use in SYBR® Green quantitative real-time PCR and have been tested in conjunction with SimpleChIP® Enzymatic Chromatin IP Kits #9002 and #9003 and ChIP-validated antibodies from Cell Signaling Technology®. Tyrosinase is an oxidative enzyme that is involved in melanin synthesis that when mutated can cause albinism.

Product Usage Information
1. Label the appropriate number of PCR tubes or PCR plates compatible with the model of real-time PCR machine to be used. PCR reactions should be performed in duplicate and should include a tube with no DNA to control for contamination, and a serial dilution of a 2% total input chromatin DNA (undiluted, 1:5, 1:25, 1:125), which is used to create a standard curve and determine amplification efficiency.2. Add 2 μl of the appropriate ChIP DNA sample to each tube or well of the PCR plate.3. Prepare a master PCR reaction mix as described below. Add enough reagents for two extra reactions to account for loss of volume. Add 18 μl of the master PCR reaction mix to each PCR reaction tube or well of the PCR plate.Reagent Volume for 1 PCR Reaction (20 μl)Nuclease-free H2O 6 μl5 μM SimpleChIP® Primers 2 μl2X SYBR® Green Reaction Mix 10 μl4. Start the following PCR reaction program:a. Initial Denaturation: 95°C for 3 min.b. Denaturation: 95°C for 15 sec.c. Anneal and Extension: Primer-specific temp. for 60 sec.d. Repeat steps b and c for a total of 40 cycles.5. Analyze quantitative PCR results using software provided with the real-time PCR machine.



Specificity/Sensitivity
Species Reactivity:
Human


Supplied in nuclease-free water at a concentration of 5 μM (each primer is at a final concentration of 5 μM). Store at -20°C.


参考图片
SimpleChIP® Human Tyrosinase Promoter Primers were tested on DNA isolated from cross-linked cells using the SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003. Real-time PCR was performed in duplicate on a serial dilution of 2% total input DNA (20 ng, 4 ng, 0.8 ng, and 0.16 ng) using a real-time PCR detection system and SYBR® Green reaction mix. The PCR amplification efficiency (E) and correlation coefficient (R2) were calculated based on the corresponding threshold cycle (CT) of each dilution sample during 40 cycles of real-time PCR (95°C denaturation for 15 sec, 65°C anneal/extension for 60 sec).
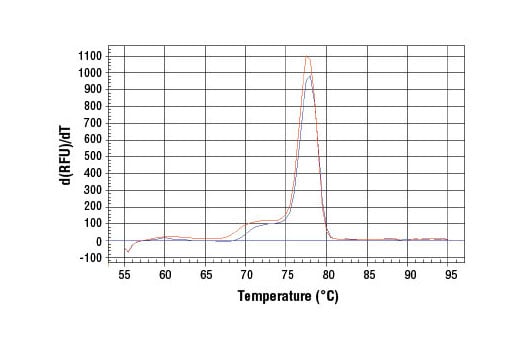
PCR product melting curves were obtained for real-time PCR reactions performed using SimpleChIP® Human Tyrosinase Promoter Primers. Data is shown for both duplicate PCR reactions using 20 ng of total DNA. The melt curve consists of 80 melt cycles, starting at 55°C with increments of 0.5°C per cycle. Each peak is formed from the degradation of a single PCR product.






 用小程序,查商品更便捷
用小程序,查商品更便捷